A University of Saskatchewan research team has published new insight into the structure of SARS-CoV-2, the virus causing COVID-19. Their findings may serve as the basis for future treatments.
Disulfide bonds, which are pairs of sulphur atoms, are known to stabilize protein structures. That these bonds are abundant in a key area of the SARS-CoV-2 virus was the foundation for the researchers’ investigation into the potential of drugs that break disulfide bonds.
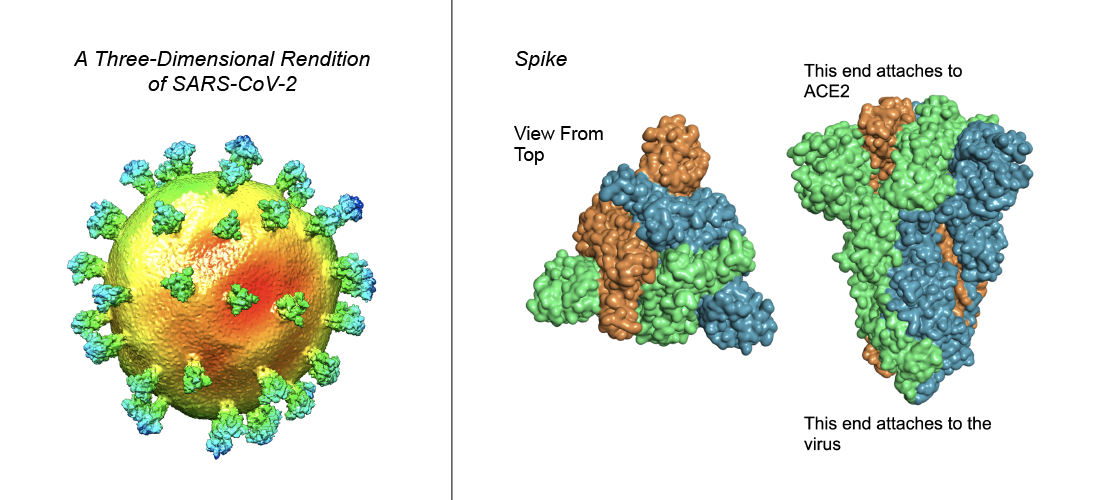
For SARS-CoV-2, like all coronaviruses, the infection of human cells begins with a protein called the envelope Spike glycoprotein. The Spike protein binds to a human cell surface receptor through a receptor-binding domain, or RBD for short. For SARS-CoV-2, the RBD binds a protein called angiotensin-converting enzyme 2, also known as ACE2.
Among the collaborators on the team were U of S professors and Canada Research Chairs Ingrid Pickering, PhD, and Graham George, PhD. In speaking to the Sheaf about the new publication, they emphasize that it was the dedication of the two USask research associates leading the research that made the project a success.
George says that it was by “complete coincidence” that Nataliya Dolgova, PhD, and Andrey Grishin, PhD, simultaneously began investigating the possibility of disrupting disulfide bonds in the RBD and ACE2 as a way to inhibit the Spike protein’s ability to infect human cells.
In summer of 2020, Dolgova began researching this potential using protein-based assays to examine the binding relationship of the Spike protein and ACE2.
Around the same time, Grishin started computer model studies to simulate the effect of breaking particular disulfide bonds in the Spike protein. Grishin also worked with U of S Vaccine and Infectious Disease Organization to conduct live virus tests using compounds known to break disulfide bonds.
After Dolgova and Grishin learned of the overlap of their projects, they chose to proceed as a team. Collaborating with researchers from across the U of S — including the departments of geological sciences, chemistry, biochemistry, microbiology and immunology, the USask Advanced Research Computing team, and VIDO — they employed various methods to procure the findings now being published in the Journal of Molecular Biology.
“[Dolgova and Grishin] had great drive and energy and it was all them that drove [the research],” George said.
The team’s paper shows that sufficiently high concentrations of disulphide-disrupting compounds can destabilize the Spike protein RBD structure and reduce the infectivity of SARS-CoV-2.
George says that a future treatment based on their findings could work for future variants of SARS-CoV-2 that result from mutations. This is because it would target both disulfide bonds in the human ACE2 protein, which will not mutate, and in the Spike protein.
“The disulfides are so integral to the structure of the Spike [protein], so important for it that there’s no way that a mutation could disrupt this very basic approach to messing up the infection process,” George said. “So, I’m encouraged by that.”
Pickering says that the support she witnessed between the scientists working on the paper was a unique aspect of the project.
“Research is often very competitive and closed off from your competitors,” Pickering said. “In other words, we’re trying to get these results and we’re trying to come up with this big find, whatever it is. What happened in this case was much more just people naturally collaborating in ways which don’t always happen.”
Since working with Dolgova and Grishin, Pickering and George say they have seen similar collaborative attitudes in their other COVID-19-related research endeavours. Pickering suggests that the openness of scientists at other institutions, beyond the U of S and Canada, to discuss overlapping research and help others in accessing resources may be a consequence of the pressing nature of the pandemic.
“I loved the way that that happened in our small project, and I hope that that’s repeated in many, many places as we seek to solve this crisis.”
A previous version of this article incorrectly stated that Nataliya Dolgova used cell-based assays to examine the binding relationship of the Spike protein and ACE2. Dolgova purified the Spike and ACE2 proteins from cells to examine them, making these protein-based assays rather than cell-based.
We apologize for these errors. If you spot any errors in an article, please email them to copy@thesheaf.com for correction.
—
Sandra LeBlanc | News Editor
Graphic: Supplied by Dr. Graham George